library(rgrass)Get started with GRASS & R: the rgrass package
The rgrass package allows us to interact with GRASS tools (and data) serving as an interface between GRASS GIS and R. The rgrass package is developed and maintained by Roger Bivand and can be found at: https://github.com/rsbivand/rgrass/. In this fast track tutorial, we will learn how to use GRASS GIS from R.
To run this tutorial locally you should have GRASS GIS 8.4+, R and, optionally, RStudio installed. You will also need to install rgrass, terra and mapview R packages and download the North Carolina sample dataset.
rgrass main functions
The main functions within rgrass are the following:
initGRASS(): starts a GRASS session from R.execGRASS(): executes GRASS commands from R.gmeta(): prints GRASS session metadata like database, project, mapset, computational region settings and CRS.read_VECT()andread_RAST(): read vector and raster maps from a GRASS project into R.write_VECT()andwrite_RAST(): write vector and raster objects from R into a GRASS project.
For further details on rgrass functionality, usage examples and data format coercion, see: https://rsbivand.github.io/rgrass/.
Basic usage: Choose your own adventure
If you are a regular R user that needs to use GRASS GIS functionality because, well, you know it rocks, rgrass has your back. For example, maybe you struggle with large raster datasets in R or you need some specific tool, like watershed delineation for a large high resolution DEM. We will show here the way to use GRASS tools within your R workflows.
On the other hand, if you already use GRASS as your geospatial data processing engine, you most likely have your spatial data within GRASS projects. You might need however to do some statistical analysis, some modelling and prediction or create publication ready visualizations in R. In such cases, you can start a GRASS session in your project from R or RStudio.
Let’s see the general basic steps and then dive into the details:
- Make sure GRASS GIS is installed.
- Open R (or RStudio)
- Load
rgrasslibrary withlibrary(rgrass) - Start a GRASS session with
initGRASS() - Use GRASS tools through
execGRASS() - Use
read_VECT(),read_RAST(),write_VECT()andwrite_RAST()to read data from and write data into GRASS database.
GRASS raster and vector maps are translated into terra’s package SpatRaster and SpatVector objects, respectively. These objects can then, within R, be easily coerced to other types of spatial objects such as simple features (sf), stars, etc.
See terra vignettes with further explanations and examples: https://rspatial.github.io/terra/.
A. Use GRASS GIS tools within your R spatial workflows
We start R or Rstudio and load the rgrass library. It will tell us that GRASS is not running, but we know that already… and that’s about to change in a moment.
In case you need to include some of the cool GRASS tools within your R workflow, the initGRASS() function allows you to create temporary GRASS projects to use GRASS tools on R objects. This is equivalent to what QGIS does when you use GRASS tools via the QGIS Processing Toolbox.
First, we will use initGRASS() to create a temporary GRASS project based on the extent, resolution and CRS of a raster or vector R object, likely the one we want to process or one that has the extent of our study area. Hence, we need to pass a reference spatial grid via the SG parameter. Then, we will write our R objects into the temporary GRASS project, run the desired processes, and export the outputs back to the R environment.
Let’s start with getting some spatial data, e.g., a raster file, into R.
library(terra)
f <- system.file("ex/elev.tif", package="terra")
r <- rast(f)
plot(r)Now, we will start GRASS GIS in a temporary folder. By specifying SG = r, the GRASS project is internally created with raster r’s object CRS (BTW, you can check that with crs(r)), extent and resolution. These latter define the GRASS computational region that will affect all raster processing, i.e., all new raster maps generated within GRASS GIS will have the same extent and resolution of the map provided. If you wish to change the computational region later on, you can use the g.region GRASS tool with execGRASS("g.region --h").
Optionally, we can specify which GRASS binary to use with gisBase. This might be useful in case we have several GRASS versions on our system. If not provided, initGRASS() will attempt to find it in default locations depending on your operating system.
# Start GRASS GIS from R
initGRASS(home = tempdir(),
SG = r,
override = TRUE)Now, we can write our SpatRaster into the GRASS temporary project.
write_RAST(r, "terra_elev")Alternatively, we can use GRASS importing tools to import common raster and vector formats. Data will be reprojected if needed.
execGRASS("r.import", input=f, output="test")Let’s check both raster maps (test and terra_elev) are indeed within the project and run the GRASS tool r.slope.aspect on one of them.
execGRASS("g.list", type = "raster")execGRASS("r.slope.aspect",
elevation = "terra_elev",
slope = "slope",
aspect = "aspect")execGRASS("g.list", type = "raster")Let’s get slope and aspect maps into R
grass_maps <- read_RAST(c("aspect", "slope"))
grass_mapsNow that the output maps are back into our R environment, we can plot them, do further analysis or write them into other raster formats, in which case we use terra::writeRaster() function.
plot(grass_maps)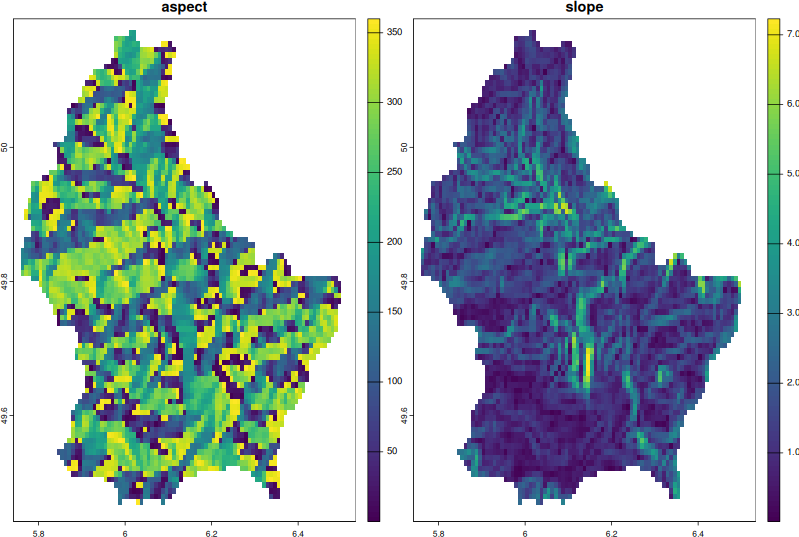
writeRaster(grass_maps, "grass_maps.tif", overwrite=TRUE)Alternatively, we can use GRASS GIS exporting tools like r.out.gdal and v.out.ogr, to directly save our outputs into common raster or vector formats, respectively.
execGRASS("r.out.gdal", input="slope", output="slope.tif", format="GTiff", flags="overwrite")B. Use R tools within GRASS GIS workflows
Let’s see an example for the case when we do our geospatial data processing within GRASS GIS and hence have all the spatial data organized within GRASS projects but we need to run some statistical analysis, modelling, prediction or visualization in R.
library(rgrass)We start GRASS GIS from within R or RStudio using the initGRASS() function. Since we want to start GRASS GIS in a specific project and mapset, we need to specify them.
# Start GRASS GIS from R
initGRASS(gisDbase = path.expand("~/grassdata/"),
location = "nc_basic_spm_grass7",
mapset = "PERMANENT",
override = TRUE,
remove_GISRC = TRUE)We can now list and read our GRASS raster and vector maps into R and do our statistical analysis, modelling and/or visualizations using other R packages. Here, we will demonstrate the use of all the main rgrass functions mentioned above.
Let’s then list our GRASS raster and vector maps:
# List GRASS raster maps
execGRASS("g.list", type="raster")# List GRASS vector maps
execGRASS("g.list", type="vector")The resulting map lists could be saved in an R object that we can subset later in case we want to import several but not all raster maps, for example. Let’s see how to do that.
# Save map list in an object
rast_list <- execGRASS("g.list", type="raster")
# Retrieve only the map list from standard output
rast_list <- attributes(rast_list)$resOut
# Import elevation and landuse
to_import <- c("elevation", "landuse") # optionally, by position: rast_list[c(3,7)]
maplist <- list()
for (i in to_import) {
maplist[[i]] <- read_RAST(i)
}
maplistRemember that raster objects will always be exported from GRASS GIS following the computational region settings. So, bear that in mind when reading into R which will hold them in memory. Vectors however will be exported in their full extent.
Let’s load the terra library to quickly display our recently imported raster maps:
library(terra)
plot(maplist$elevation)Optionally, we could stack our two SpatRaster objects together and plot them together:
rstack <- rast(maplist)
plot(rstack)Let’s create a boxplot of elevation per land class.
boxplot(rstack$elevation, rstack$landuse, maxcell=50000)Let’s import a vector map, too, and explore its attributes.
census <- read_VECT("census")
head(census)summary(census$TOTAL_POP)plot(census, "P25_TO_34", type="interval", breaks=5, plg=list(x="topright"))Let’s do some interactive visualization with mapview.
library(mapview)
mapview(rstack$elevation) + censusWe highly recommend you to check the tmap package to make really appealing and publication ready maps.
To exemplify the use of write_* functions, let’s do a simple operation with the landuse raster map. We will apply a custom function that makes NULL all values less than 4.
result <- app(rstack$landuse, fun=function(x){ x[x < 4] <- NA; return(x)} )
plot(result)To use this new raster in GRASS GIS, for example as an input to a GRASS tool, we need to call write_RAST function:
write_RAST(result, "result_from_R", overwrite = TRUE)The new raster is now written as a GRASS raster and can be listed:
execGRASS("g.list", parameters = list(type="raster", pattern="result*"))Finally, there is yet another way in which you can use GRASS and R together, and it involves calling R from the GRASS terminal. In this way, rgrass will read all GRASS session environmental variables, and you won’t need to use initGRASS(). It goes more or less like this:
- Open GRASS GIS
- Type
Rorrstudio &in the GRASS terminal - Load
rgrasslibrary withlibrary(rgrass) - Use
read_VECT(),read_RAST()to read data from GRASS GIS into R - Do your analysis or plotting in R
- Write data (back) to GRASS database with
write_VECT()andwrite_RAST() - Quit R
quit()and get back to GRASS terminal.
Starting GRASS GIS...
__________ ___ __________ _______________
/ ____/ __ \/ | / ___/ ___/ / ____/ _/ ___/
/ / __/ /_/ / /| | \__ \\_ \ / / __ / / \__ \
/ /_/ / _, _/ ___ |___/ /__/ / / /_/ // / ___/ /
\____/_/ |_/_/ |_/____/____/ \____/___//____/
Welcome to GRASS GIS 8.4.0
GRASS GIS homepage: https://grass.osgeo.org
This version running through: Bash Shell (/bin/bash)
Help is available with the command: g.manual -i
See the licence terms with: g.version -c
See citation options with: g.version -x
If required, restart the GUI with: g.gui wxpython
When ready to quit enter: exit
Launching <wxpython> GUI in the background, please wait...
[Raster MASK present]
GRASS nc_basic_spm_grass7/PERMANENT:~ > R
R version 4.3.1 (2023-06-16) -- "Beagle Scouts"
Copyright (C) 2023 The R Foundation for Statistical Computing
Platform: x86_64-pc-linux-gnu (64-bit)
R is free software and comes with ABSOLUTELY NO WARRANTY.
You are welcome to redistribute it under certain conditions.
Type 'license()' or 'licence()' for distribution details.
Natural language support but running in an English locale
R is a collaborative project with many contributors.
Type 'contributors()' for more information and
'citation()' on how to cite R or R packages in publications.
Type 'demo()' for some demos, 'help()' for on-line help, or
'help.start()' for an HTML browser interface to help.
Type 'q()' to quit R.
> library(rgrass)
GRASS GIS interface loaded with GRASS version: GRASS 8.4.0 (2024)
and location: nc_basic_spm_grass7
> Enjoy!
References
- Bivand R (2024). rgrass: Interface Between ‘GRASS’ Geographical Information System and ‘R’. R package version 0.4-1, https://CRAN.R-project.org/package=rgrass.
The development of this tutorial was funded by the US National Science Foundation (NSF), award 2303651.